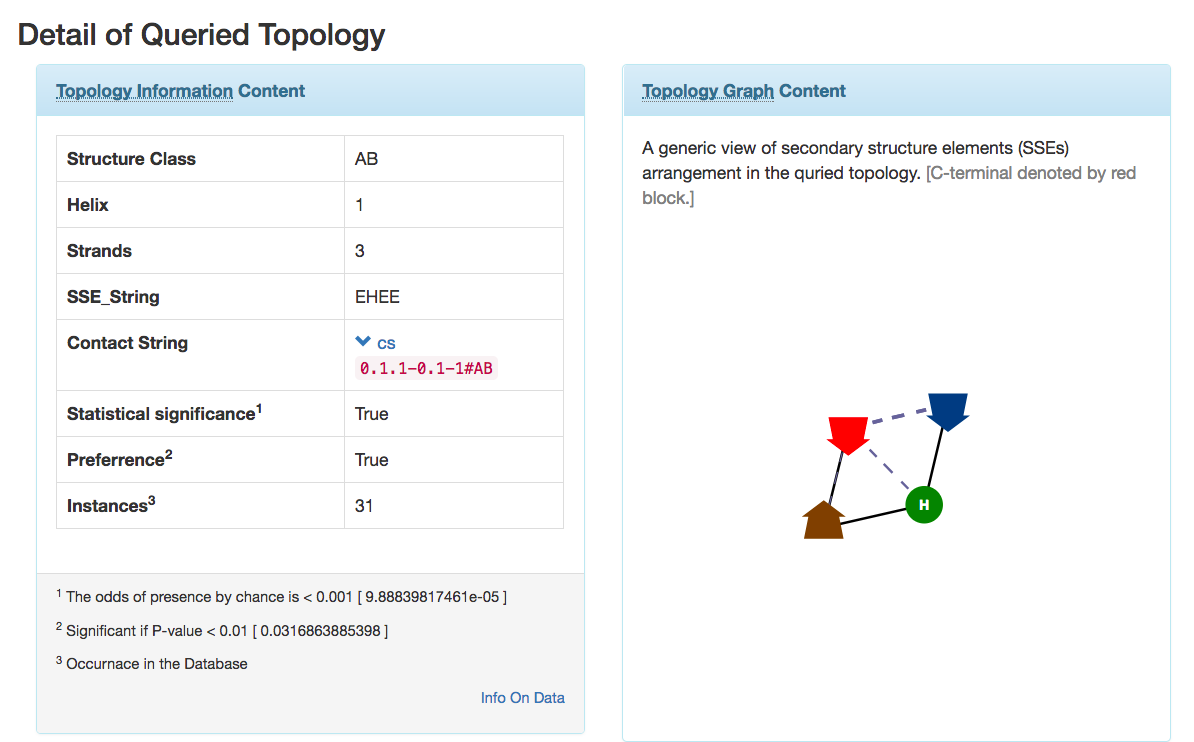
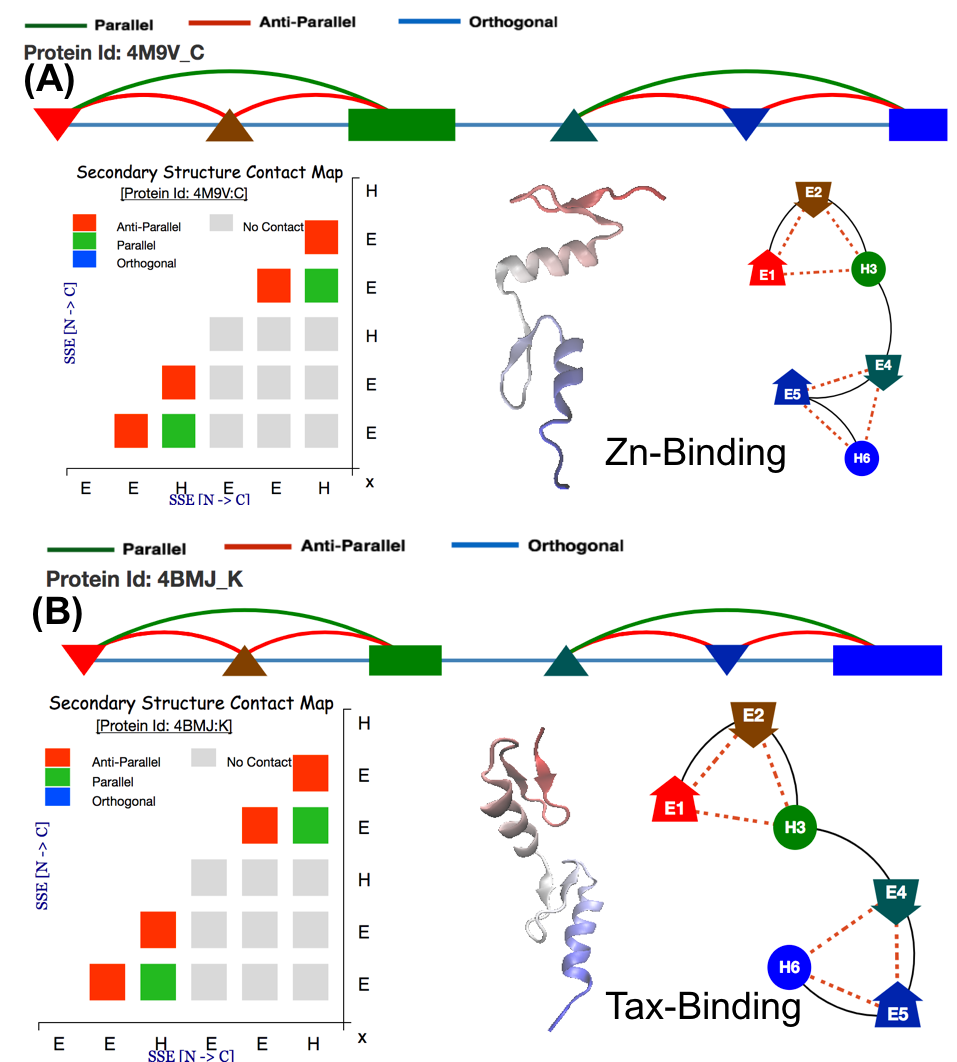
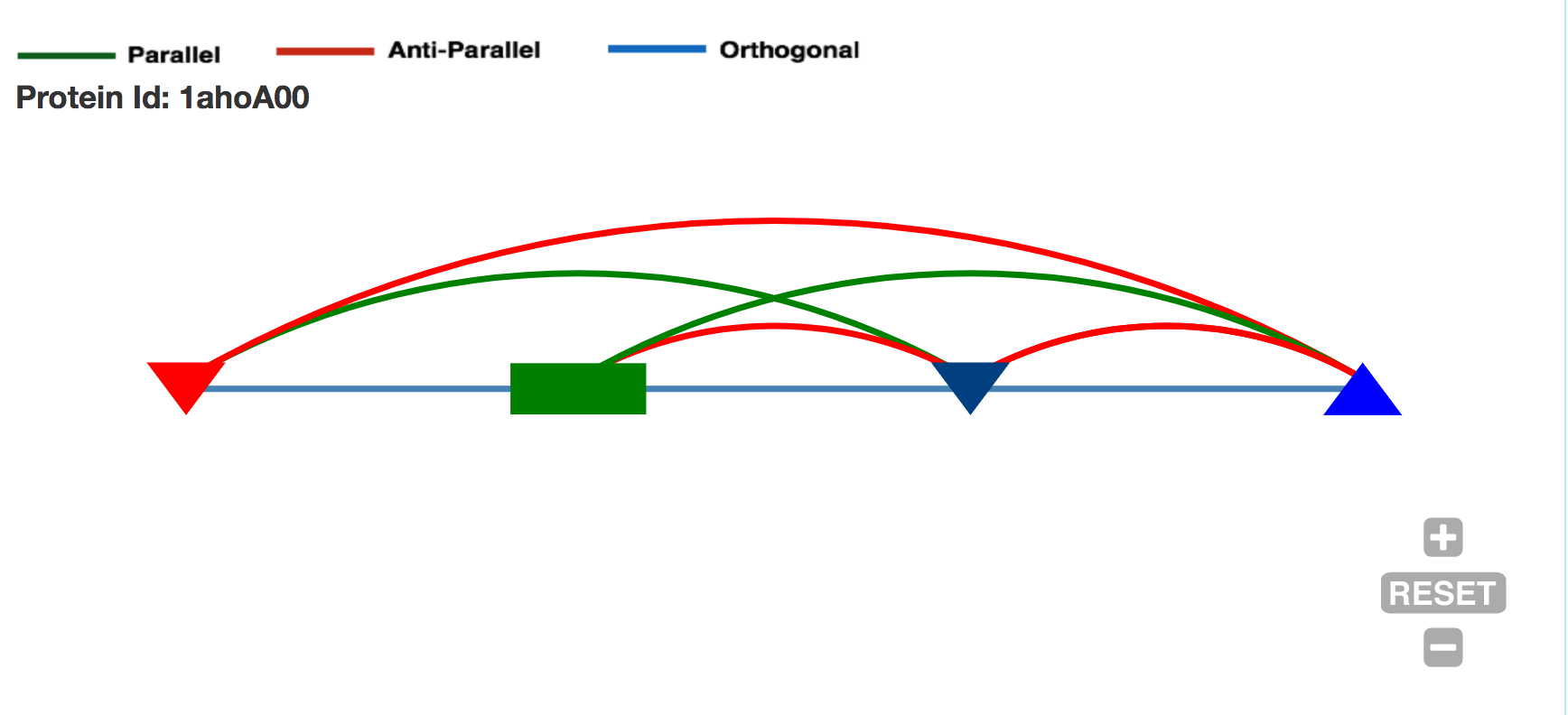
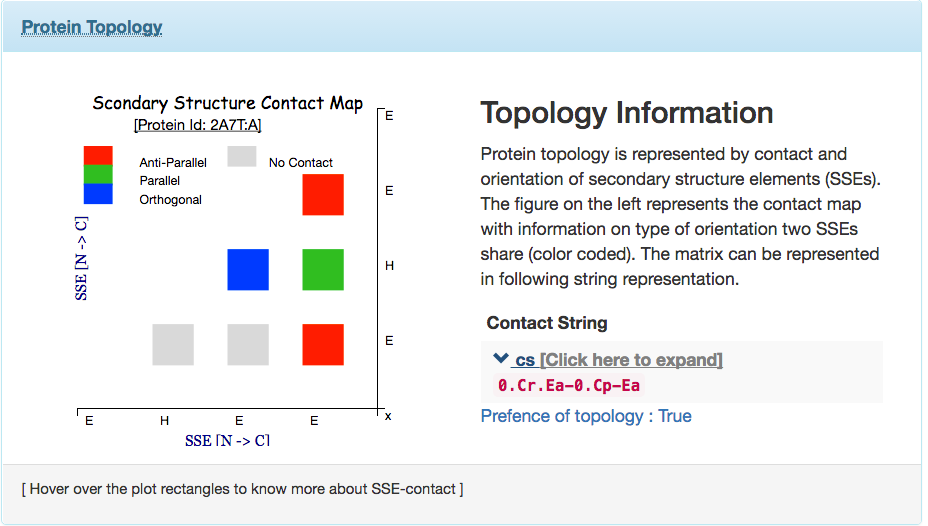
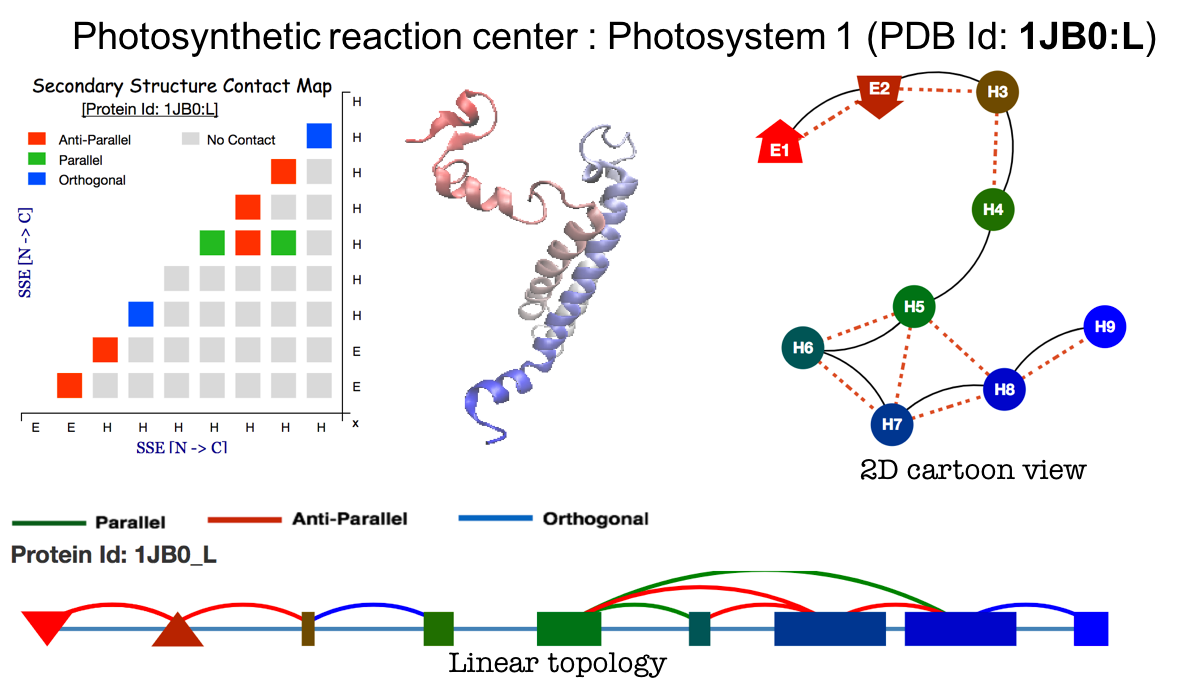
Contact String the Topology representation : Given a protein structure, ProLego enumerates constituent secondary structures and their arrangements. This is represented as a unique 1D string.
Generate topology visualization:
-
A 2D topology representation
-
A 1D or Linear topology representation
where secondary structures will be present as per their chain
connectivity N to C and contact between SSEs will be assigned by
different arcs connecting them.
Topology Classifier:
Comparing the obtained “contact string” with studied datasets, ProLego
assigns frequency of occurrence status (prevalent/non-prevalent) for the
corresponding topology. If the topology is not available in the dataset
it should be able to update database on the fly.
Module Analysis: Given a contact String ProLego server can extract all the possible modules and searches similar proteins in the database.
Contact Map: Plots secondary structure contact map given a protein structure